 Author
Author  Correspondence author
Correspondence author
International Journal of Molecular Medical Science, 2024, Vol. 14, No. 1 doi: 10.5376/ijmms.2024.14.0005
Received: 02 Feb., 2024 Accepted: 05 Mar., 2024 Published: 19 Mar., 2024
Wang W., 2024, Studying the dynamic changes of the immune system through single-cell omics, International Journal of Molecular Medical Science, 14(1): 29-41 (doi: 10.5376/ijmms.2024.14.0005)
The rapid development of single-cell omics technologies has provided a new perspective for immunology research, enabling scientists to explore the dynamic changes of the immune system at unprecedented resolution. This study reviews the application cases, challenges, and future development directions of single-cell omics in immune system research. This study introduces how single-cell omics technologies reveal the heterogeneity and functional diversity of immune cells in infectious diseases, autoimmune diseases, and tumor microenvironments. Single-cell RNA sequencing technology and single-cell ATAC sequencing technology enable scientists to identify immune cell subsets and their gene expression patterns under specific disease conditions, thereby deeply understanding how pathogens affect the functional state of immune cells and how the immune system adjusts its response strategy to fight against diseases. These advancements will help comprehensively understand the state and function of cells, as well as their dynamic changes in health and disease states. At the same time, it emphasizes the important impact of single-cell omics technology on disease treatment, especially personalized medicine and precision immunotherapy. By analyzing the immune cell status of specific patients, personalized treatment plans can be customized to improve the effectiveness and safety of treatment. Single-cell omics technology provides powerful new tools for the study of the immune system, but it also brings new challenges. With the continuous advancement of technology and the development of new methods, it is expected to overcome these challenges, deeply explore the mysteries of the immune system, and provide deeper insights and more effective solutions for immunology research and clinical treatment. In the future, single-cell omics will continue to play a key role in revealing the internal complex mechanisms of the immune system and promoting personalized medicine, providing more precise treatment targets and methods, thereby greatly improving treatment effects and patient quality of life.
The complexity and dynamics of the immune system have long been central topics in biomedical research. While traditional population-level approaches have made progress in revealing overall trends in immune response, they have not been able to resolve the subtle differences between cells and the unique behavior of individual cells. With the rapid development of single-cell omics technology, especially single-cell transcriptome sequencing technology, it is possible to study the dynamic changes of the immune system with unprecedented resolution, thereby revealing the heterogeneity of immune cells, the transformation of cell states, and their specific functions in health and disease (Stubbington et al., 2017).
Single-cell techniques not only provide the possibility to identify and classify new immune cell subtypes, but also reveal the complex regulatory networks and signaling pathways inside cells. These cell-level insights are critical to understanding how the immune system responds to various external stimuli, both in immune regulation and during immune-mediated disease development (Vegh and Haniffa, 2018). In addition, single-cell technology has shown great potential in revealing the clonality and diversity of immune cells such as T cells and B cells, providing a new perspective for vaccine development and immunotherapy.
Although single-cell omics technology has brought breakthroughs in immunology research, the analysis and interpretation of data still face challenges. The processing of high-dimensional data, precise identification of cell states, and dynamic modeling of complex biological processes require researchers to develop new computational methods and analytical tools (Tanay and Regev, 2017). This study reviews the use of single-cell omics techniques in studying dynamic changes in the immune system, explores key biological findings revealed by these techniques, and discusses strategies to overcome current challenges, as well as the potential impact of these techniques on future immunology research and clinical applications.
1 Overview of Monocytomic Techniques
1.1 Single-cell RNA sequencing (scRNA-seq)
With the continuous progress of science and technology, people's understanding of life science is gradually deepening. Among them, single-cell RNA sequencing technology, with its unique advantages, has revealed the mystery of the inner world of cells for everyone. This technology enables precise sequencing of RNA molecules in individual cells to map the cell's gene expression patterns, providing a deeper understanding of the complexity and diversity of cells (Figure 1).
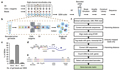 Figure 1 The scRNA-seq workflow |
Islam et al. (2013) proposed that the core of single-cell RNA sequencing technology lies in its high sensitivity and high resolution. Traditional RNA sequencing methods typically process large numbers of cells in batches, making it difficult to capture small differences between cells. Single-cell RNA sequencing, on the other hand, is able to analyze individual cells and see the gene expression of each cell in detail. This allows researchers to more accurately identify different cell subpopulations and further understand inter-cell heterogeneity. Single-cell RNA sequencing technology will play an increasingly important role in the field of life sciences and make greater contributions to human health and development.
Kashima et al. (2020) believe that single-cell RNA sequencing technology is expected to play a greater role in disease diagnosis, treatment and new drug research and development. By analyzing individual cells in a patient's body, doctors can develop more precise treatment plans and improve the cure rate of the disease. At the same time, the technology also helps to discover new drug targets and develop more effective drugs, making greater contributions to human health. As an important breakthrough in the field of life science, single-cell RNA sequencing technology has revealed the mystery of the inner world of cells, and has also enabled researchers to have a deeper understanding of the complexity and diversity of cells, injecting new vitality into the field of life science research. With the continuous development and improvement of the technology, single-cell RNA sequencing technology will play a more important role in the future and make greater contributions to human health and development.
1.2 Single-cell ATAC sequencing (scATAC-seq)
The scATAC-seq technique is used to analyze the accessibility of chromatin in individual cells to infer the open status of gene regulatory regions. This is essential for understanding the mechanisms of cell type-specific gene expression regulation.
Conklin et al. (2022) proposed that the development of TEA-seq technology is a three-mode sequencing method that can simultaneously measure the transcriptome (scRNA-seq), epitopes, and chromatin accessibility (scATAC-seq) of a single cell, providing a new tool for cell type-specific gene regulation and expression.
According to Patruno et al. (2023), the launch of ArchR software package provides a rapid and comprehensive analysis of single-cell chromatin accessibility data, including single-cell clustering, cell type identification, DNA element and gene link analysis, etc., which greatly accelerates the understanding of gene regulation (Granja et al., 2021).
Schep et al. (2017) suggest that chromVAR, an R package for analyzing sparse single-cell ATAC-seq data, is able to accurately cluster scATAC-seq profiles and identify known and new moths associated with changes in chromatin accessibility by estimating accessibility gains or losses within peaks that share the same moths or notes. These studies not only expand the understanding of chromatin accessibility and gene regulation mechanisms at the single-cell level, but also provide new tools and analytical frameworks for future research.
1.3 Single cell mass spectrometry (CyTOF)
In cell biology research, a technology called CyTOF (Cytometry by Time-Of-Flight), with its unique advantages, is gradually becoming an important tool to study the phenotype and function of immune cells. The core of CyTOF technology is its ability to label proteins on the cell surface and inside the cell, thereby quantifying tens to hundreds of proteins at the single-cell level. This property allows researchers to gain insight into the complexity and diversity of cells in the microscopic world, which in turn reveals the nature of life activities.
According to Wang and Navin's (2015) study, CyTOF has higher parametric capacity and lower background noise compared with traditional flow cytometry. Although flow cytometry can also analyze proteins on the cell surface to a certain extent, due to the limitations of its working principle, it is often difficult to analyze multiple proteins at the same time, and the background noise is high, affecting the accuracy of the results. CyTOF technology, through its unique time-flight principle, can significantly increase the parameter capacity of analysis while maintaining high sensitivity, making it possible to analyze tens or even hundreds of proteins at the same time in a single experiment.
Proserpio and Mahata (2015) proposed that the application of CyTOF technology is even more convenient in the study of immune cells. As an important force of human body to resist the invasion of foreign pathogens, the diversity of immune cells' phenotypes and functions is crucial for maintaining body homeostasis. CyTOF technology can accurately quantify the expression levels of receptors, ligands and intracellular signaling molecules on the surface of immune cells, thereby revealing the mechanism of action of different immune cell subsets in the occurrence and development of diseases. Researchers can use CyTOF technology to analyze the phenotype and function of immune cells in the tumor microenvironment. By comparing the immune cells in tumor tissue and healthy tissue, researchers can find the changes in the type, number and functional status of tumor-infiltrating immune cells, providing new ideas and methods for tumor immunotherapy.
1.4 Single cell imaging technology
Chen et al. (2015) believe that single-cell imaging, as an important tool for modern biological research, covers both optical microscopy and electron microscopy, providing intuitive information on cell morphology, location and interactions. These techniques allow scientists to delve deeper into the complex mechanisms inside cells, opening up new horizons for biomedical research. Optical microscopy is an important part of single cell imaging. By utilizing different wavelengths of light and special fluorescent markers, light microscopes are able to reveal the fine structure and function inside cells. Fluorescence microscopy allows specific protein or nucleic acid molecules to be clearly visible under the microscope by exciting fluorescent molecules within cells. This technique provides researchers with a visual picture of the dynamic processes inside cells, helping to understand key biological processes such as cell signal transduction, gene expression and protein interactions.
Gomes et al. (2019) suggest that by replacing light with electron beams, electron microscopes are able to penetrate the fine structures inside cells, revealing more details about cell structure and function. This allowed the researchers to observe information such as the ultrastructure of organelles, the arrangement of the cytoskeleton, and the distribution of molecules on the cell membrane. Single cell imaging technology plays an irreplaceable role in biomedical research with its unique advantages. By visually demonstrating cell morphology, location, and interactions, these technologies not only provide insight into the complex mechanisms inside cells, but also provide new ideas and methods for disease diagnosis and treatment. With the continuous advancement of technology, single-cell imaging technology is expected to reveal more microscopic mysteries about the cellular world in the future.
1.5 Advantages and limitations of technology
Single-cell omics technology provides a direction for in-depth insight into the microscopic world of biological systems. In the past, technical limitations have made it difficult for scientists to conduct detailed analyses of individual cells. However, with monocytomics, a comprehensive genome, transcriptome, and proteome level study of individual cells can be performed to gain a more accurate understanding of cell properties and functions.
Kolodziejczyk et al. (2015) believe that in the field of immunology research, monocytomics technology has shown great potential. It helps reveal the complexity of immune responses, including identifying new immune cell subtypes, understanding the dynamics of immune cells in healthy and disease states, and how immune cells interact in different tissue microenvironments. These studies not only contribute to a deeper understanding of how the immune system works, but also provide new ideas and methods for disease prevention and treatment. However, as with any technology, there are certain limitations to single-cell omics technology. The cost of this technology is relatively high, including inputs such as experimental equipment, reagents and data analysis. In addition, the data processing and analysis process is also very complex and requires specialized computational biology support. This, to some extent, limits the progress in the wide application and popularization of the technology.
Butler et al. (2018) proposed that with the continuous advancement of technology and the gradual reduction of cost, single-cell omics technology is expected to play a greater role in the future. For example, by further optimizing experimental methods and data analysis algorithms, the accuracy and efficiency of research can be improved, thus making better use of this technology to explore the mysteries of life sciences. Single-cell omics technology brings new opportunities and challenges to biological research with its unique advantages. Although there are still some limitations, with the continuous improvement and development of the technology, it is believed that this technology will play a more important role in the future and make greater contributions to the progress of human health.
2 Study on Dynamic Changes of Immune System
2.1 Heterogeneity and functional diversity of immune cells
Immune cells exhibit a high degree of heterogeneity and functional diversity, which is key to the immune system's ability to respond to a wide variety of threats. Single-cell omics techniques are able to study this heterogeneity at the level of individual cells, revealing subtle differences even in seemingly homogeneous populations of cells. These differences may reflect the characteristics of cells at different stages of development, or different responses to environmental signals.
Levine et al. (2017) submitted in their study that multiple T cell subsets, including different types of helper T cells (Th1, Th2, Th17, etc.) and regulatory T cells (TreGs), can be identified through single-cell RNA sequencing technology. These cells play different roles in the immune response, from promoting inflammatory responses to maintaining immune tolerance, and the diversity of their functions is achieved through different gene expression programs. This diversity of cell types and functional flexibility allows the immune system to respond to a wide variety of threats. These techniques study gene expression, protein interactions and metabolic pathways at the level of individual cells, revealing subtle differences even in seemingly homogeneous populations of cells. These differences may reflect the characteristics of cells at different stages of development, or different responses to environmental signals.
Giladi and Amit (2018) believe that in addition to the heterogeneity of immune cells, the dynamic changes of the immune system are also reflected in the interactions between immune cells and microorganisms. There are a large number of microorganisms in the body, including bacteria, fungi and viruses. These microbes form a delicate balance with the body's immune system. On the one hand, microbes boost resistance by stimulating the immune system; The immune system, on the other hand, maintains this balance by eliminating and limiting the growth of microorganisms. However, when this balance is upset, such as when the microbes are too numerous or too toxic, the immune system initiates a more intense response to clear out these threats. This reaction can cause side effects such as tissue damage and inflammation, but it also provides the body with necessary protection.
2.2 Cell dynamics during immune response
Immune response is a highly dynamic process involving activation, proliferation, migration and functional execution of a variety of immune cells. Monocytomics techniques reveal cell dynamics in this process, including how cells change from a resting state to an active state, how they migrate between different parts of the body, and how they interact with other cells. For example, when a pathogen invades, dendritic cells capture and process antigens, then migrate to the lymph nodes to activate T cells. Changes in the phenotypes of dendritic cells and T cells were observed during this process, as well as the interactions between them.
Francisco et al. (2010) argue that monocytomic techniques reveal how cells transition from a resting state to an active state, how they migrate between different parts of the body, and how they interact with other cells. Using this technique, we can observe the real-time dynamics of immune cells in the body with unprecedented precision and depth. In the case of a pathogen invasion, the immune system immediately activates its defense mechanisms. Dendritic cells play a crucial role in this process. They are able to quickly capture and process invading antigens and then pass this information on to other immune cells. The dendritic cells then migrate to the lymph nodes, activating the T cells and triggering a stronger immune response.
Wills and Mead (2015) mentioned in their study that changes in dendritic cell and T cell phenotypes during this process can be clearly observed through single-cell tracking techniques. After capturing antigens, dendritic cells undergo a series of changes in their surface molecules that allow them to better interact with other immune cells. At the same time, after being activated, T cells will also undergo a series of proliferation and differentiation to adapt to the need to fight pathogens. These research results not only provide a deeper understanding of the mechanism of immune response, but also provide new ideas and methods for future immunotherapy. By manipulating the dynamic processes of these immune cells, it may be possible to prevent and treat various diseases more effectively.
2.3 Cellular basis of inflammatory response and immune tolerance
The immune system is a delicate and complex network that maintains a delicate balance between inflammatory response and immune tolerance. This balance is essential to ensure effective removal of pathogens and to protect the body from infection, while also avoiding damage to one's own tissues from unwanted immune responses. In recent years, with the rapid development of single-cell omics techniques, a deeper understanding of the cellular basis behind this balance has been gained.
Gomes et al. (2019) mentioned that regulatory T cells (TREGs) are one of the key cell types in maintaining immune tolerance. They act as a "brake" to ensure that the immune response does not overshoot, preventing autoimmune diseases from developing. Treg cells, however, are not monolithic and contain a variety of subtypes that exhibit different functions in different tissues and inflammatory conditions. Through single-cell RNA sequencing, the researchers found that these subtypes not only differed in their expression profiles, but also were distinctive in regulating the immune response. For example, some Treg cell subtypes may be better at inhibiting the production of inflammatory mediators, while others may be better at promoting tissue repair and regeneration. This diversity and functionality allows Treg cells to flexibly adjust their immune regulatory strategies according to different inflammatory environments and tissue needs.
Duhen et al. (2012) mentioned that monocytomics technology provides a new perspective, allowing researchers to better understand how the immune system maintains a balance between inflammatory response and immune tolerance. This not only contributes to a better understanding of the pathogenesis of immune-related diseases, but also provides an important theoretical basis for the development of new immunotherapies. With the continuous progress of technology and in-depth research, future immunological research will be more accurate and efficient, and bring greater benefits to human health and well-being.
2.4 Relationship between immune escape mechanism and disease progression
The progression of disease, especially cancer, is often accompanied by a phenomenon known as immune escape. Immune escape, in short, means that diseased cells evade the surveillance and attack of the immune system through a series of strategies. The presence of this mechanism allows tumor cells to continue to grow and spread in the body, thus exacerbating the disease process.
Duhen et al. (2012) explored in detail the state of immune cells in the tumor microenvironment, providing valuable data for understanding how tumors evade immune responses. By using single-cell omics techniques to precisely analyze the phenotype and function of various immune cells in the tumor microenvironment, it has revealed how tumors evade immune system attack by changing the characteristics of immune cells. Tumor cells inhibit T cell activity by expressing a series of immune checkpoint proteins, such as PD-L1. These immune checkpoint proteins act as barriers that prevent T cells from effectively recognizing and attacking tumor cells. The existence of this mechanism makes tumor cells "invisible" under the eyelids of the immune system, so as to escape the immune response.
Wimmers et al. (2021) mentioned that in addition to immune checkpoint proteins such as PD-L1, tumor cells may also evade immune system attack in other ways. For example, tumor cells can suppress immune cell activity by secreting a range of immunosuppressive factors, such as transforming growth factor β(TGF-β) and interleukin-10 (IL-10). These immunosuppressive factors can interfere with immune cell signaling and function execution, making the immune system unable to effectively recognize and attack tumor cells. It is worth mentioning that the application of single-cell sequencing technology provides strong support for in-depth understanding of immune escape mechanism. Through single-cell sequencing technology, researchers can analyze the genome, transcriptome and epigenome of individual immune cells at multiple levels, thereby revealing the complex changes of immune cells in the tumor microenvironment. This technique can more accurately identify the immune escape strategy of tumor cells, and also provide important basis for the development of new immunotherapy strategies.
3 Application Cases of Monocytomics in Immune System Research
3.1 Immune cell response in infectious diseases
As an important defense line in the human body, the immune system plays a vital role in resisting the invasion of various pathogens. In recent years, with the continuous development of monocytomics technology, its application in immune system research is becoming more and more extensive. This paper will discuss the application of monocytomics in the study of immune system by taking the immune cell response in infectious diseases as an example.
Kazer et al. (2020) mentioned in their study that single-cell omics technology provides a powerful tool for revealing immune cell response mechanisms to various pathogens. Taking HIV and influenza virus infections as examples, researchers have been able to deeply analyze the dynamic changes of immune cells during infection through single-cell RNA sequencing technology. These changes involve not only the switching of cell activation states, but also the interactions between immune cells and other cell types. Through single-cell sequencing analysis, the researchers were able to paint a fine picture of how pathogens affect the functional state of immune cells and how the immune system fights infection by adjusting its response strategy.
Butler et al. (2018) believe that in addition to single-cell RNA sequencing technology, single-cell ATAC sequencing also plays an important role in the process of pathogen infection. This technique is able to reveal changes in the chromatin state of immune cells, thus providing important clues to understand the molecular mechanisms of immune memory. Immune memory refers to the ability of the immune system to respond quickly and effectively to another infection after it has experienced one. Through single-cell ATAC sequencing, researchers were able to gain insight into the regulatory mechanisms of gene expression in immune cells during infection, providing theoretical support for the development of novel vaccines and treatments.
Kolodziejczyk et al. (2015) believe that single-cell omics technology still faces some challenges in practical application. For example, the sample acquisition and processing process may have an impact on the activity of immune cells, thus affecting the accuracy of the results. In addition, the analysis and interpretation of monocytomic data requires a high level of specialized knowledge and skills. However, with the continuous advancement of technology and optimization of methods, these problems are expected to be solved.
3.2 Immune cell heterogeneity in autoimmune diseases
Autoimmune diseases are a class of complex diseases caused by the immune system mistakenly recognizing and attacking its own tissues. In recent years, with the rapid development of monocytomics technology, people have a deeper understanding of the heterogeneity of immune cells in autoimmune diseases. This heterogeneity is not only reflected in the diversity of cell types, functions and expression profiles, but also in the dynamic changes in disease development and the differences in treatment response.
Wimmers et al. (2021) mentioned in their study that monocytomic technology reveals the extremely high heterogeneity of immune cells in patients with autoimmune diseases through high-resolution analysis of gene expression profiles, proteomics, metabolomics and other information of individual immune cells. This heterogeneity is manifested not only between different cell types, such as T cells, B cells, macrophages, etc., but also between different subpopulations within the same cell type. These subgroups play different roles in the onset, development and outcome of the disease, some may promote inflammatory responses, while others may suppress immune responses or participate in tissue repair.
Taking systemic lupus erythematosus (SLE) and rheumatoid arthritis (RA) as examples, Gomes et al. (2019) found through single-cell RNA sequencing and TCR/BCR sequencing analysis that specific T cell and B cell subpopulations are present in these diseases, and these subpopulations show characteristic activity patterns in the pathological process of the disease. For example, certain T cell subpopulations may be overactivated and secrete large amounts of inflammatory factors, leading to tissue damage; Other T cell subpopulations may play a regulatory role in suppressing excessive immune responses. Similarly, specific subpopulations of B cells may produce antibodies against their own antigens and participate in disease progression; Other subpopulations of B cells may produce protective antibodies that help the body fight off disease.
3.3 Composition and function of immune cells in tumor microenvironment
The tumor microenvironment is an extremely complex and dynamic ecosystem that includes tumor cells, immune cells, stromal cells, and a variety of growth factors and signaling molecules. In this ecosystem, immune cells play a crucial role, not only interacting with tumor cells, but also influencing tumor growth, invasion, and metastasis. With the rapid development of single-cell omics techniques, researchers are able to reveal the composition and function of immune cells in the tumor microenvironment with unprecedented resolution (Figure 2).
 Figure 2 Tumor-stroma metabolic communications in the TME |
Chen et al. (2015) mentioned in their study that with the advent of single-cell RNA sequencing technology, transcriptome analysis of a single cell can be performed to identify multiple immune cell subsets in the tumor microenvironment. These subpopulations include tumor-specific T cells, regulatory T cells, and tumor-associated macrophages, which play an important role in tumor immune response. For example, tumor-specific T cells are able to recognize and kill tumor cells, while regulatory T cells may suppress anti-tumor immune responses and promote tumor growth. In addition, tumor-associated macrophages also play an important role in the tumor microenvironment, which can influence tumor growth and invasion by secreting a variety of growth factors and signaling molecules.
As mentioned in the study of Yuan Shiyang et al. (2017), single-cell sequencing technology can also reveal the complexity of immune cell expression programs in the tumor microenvironment. Each immune cell subpopulation has its own unique gene expression profile, which reflects not only the identity and function of the cell, but also the microenvironment in which the cell resides and the signaling stimuli it experiences. Therefore, single-cell sequencing technology allows for a deeper understanding of the behavior and function of immune cells in the tumor microenvironment and how they interact with tumor cells to influence tumor growth and invasion.
Wimmers et al. (2021) mentioned that single-cell omics technology brings new opportunities and challenges for tumor immunology research. This technique enables a deeper understanding of immune cell composition and function in the tumor microenvironment, leading to the discovery of new immunotherapeutic targets and approaches. For example, single-cell sequencing was used to analyze peripheral blood mononuclear cells from glioblastoma patients treated with CMV pp65-LAMP RNA pulsed dendritic cell vaccine. They found that vaccine treatment elicits dynamic changes in immune cell subpopulations and functional states, which provides important clues for developing more effective immunotherapy strategies. The composition and function of immune cells in tumor microenvironment is an important field in tumor immunology. With the continuous development of single-cell omics technology, it is possible to better understand this complex ecosystem, which will bring new breakthroughs in tumor immunotherapy.
3.4 Individual differences in vaccine reactivity
Chen et al. (2015) believe that vaccines, as a key means to prevent infectious diseases, are widely used and important. However, there are significant differences in the reactivity of different individuals to vaccines, which not only affects the effectiveness of vaccination, but also may lead to adverse reactions. In order to gain a deeper understanding of this reactivity difference, the researchers began to use single-cell omics techniques to explore the mechanisms behind it. Single-cell omics techniques, in particular single-cell RNA sequencing, provide a powerful tool to precisely reveal how immune cells react after vaccination. By collecting blood samples at different time points after vaccination, researchers can analyze these samples for single-cell RNA sequencing to see which cells are activated and how their gene expression changes.
Peng Yiman et al. (2023) believe that single-cell RNA sequencing analysis can help to understand why some people have a stronger response to vaccines while others have a weaker response, and provide an important basis for designing more effective vaccines and personalized immunization strategies. By comparing the immune responses of different individuals, a number of key genes or pathways can be identified that may be important factors affecting vaccine reactivity. In addition to revealing differences in immune reactivity, monocytomic techniques provide insight into the heterogeneity of immune cells. Immune cells play a vital role in the human body, they are not only involved in immune defense, but also closely related to the occurrence and development of many immune-related diseases. Subtle differences between different immune cell types in health and disease states can be found through single cell omics analysis, and these differences may be key to the differences in immune response.
Wimmers et al. (2021) analyzed individual immune cells following influenza vaccination by single-cell sequencing and ATAC sequencing. They found that vaccines can induce long-lasting changes in the epigenetic and transcriptional states of immune cells, which provides important clues to understanding the molecular mechanisms of immune memory. This research not only reveals how vaccines affect the internal state of immune cells, but also provides a powerful tool for developing new treatments and vaccines. By delving into individual differences in vaccine reactivity, we can better understand the complexity of the immune system and potentially lead to new breakthroughs in personalized medicine and immunotherapy. Single-cell omics technology provides strong support for the study of individual differences in vaccine reactivity.
4 Challenges and Prospects
4.1 Technical challenges
The starting point of monocytomic research is to obtain high quality single cell suspensions. In this process, sample acquisition, processing, and preservation methods are crucial, as they directly affect the state of the cell, which in turn affects the accuracy of the experimental results. If the cells are damaged or changed during harvesting or handling, the results of subsequent experiments can be skewed, leading to a misunderstanding of the internal mechanisms of the cells. At present, how to effectively isolate individual cells from solid tissues while maintaining their original state remains a technical challenge. This requires continuous exploration and innovation by researchers to seek more refined and efficient sample preparation methods. For example, some research teams are trying to use microfluidic technology or laser capture micro-cutting technology to achieve more accurate and efficient single cell separation.
Wills and Mead (2015) mentioned in their study that the huge amount of data generated by single-cell omics experiments requires researchers to have highly complex computational methods in data analysis and interpretation. Extracting meaningful biological information from data from hundreds of thousands or even millions of cells is an extremely difficult task. This requires not only efficient algorithms, but also a lot of computing resources to support it. The high dimensional and sparse nature of single-cell data also creates additional challenges for data analysis. High dimensionality means that the data contains a large number of characteristic variables, while sparsity means that each cell may only express a small percentage of those genes. How to accurately classify, judge the state and interpret the function of cells in such data background is the key problem that needs to be solved.
Yost et al. (2019) mentioned in their study that in order to address these challenges, researchers are constantly developing new data analysis methods and tools. For example, some teams are trying to leverage algorithms such as machine learning or deep learning to improve the efficiency and accuracy of data analysis. At the same time, several novel sequencing techniques are being developed to improve the resolution and data quality of single-cell omics experiments. Although monocytomics faces many technical challenges, with the continuous efforts and innovations of researchers, we believe that these problems will eventually be solved.
4.2 Biological challenges
The state of cells is transient, and they make rapid and precise adjustments in response to subtle changes in the external environment. This dynamic nature makes it extremely difficult to capture the complete picture of a cell's state. Although single-cell omics techniques can provide researchers with accurate snapshots of cells at a moment in time, how to connect these isolated snapshots to form a continuous dynamic trajectory of cell behavior has become a huge challenge. This requires researchers not only to have high-precision technical means, but also to have deep biological insight and innovative thinking.
Yost et al. (2019) argue that even if the states of cells can be accurately captured, understanding what those states mean biologically is a difficult task. The state of cells does not exist in isolation; they interact intricately with surrounding cells, tissues, organs, and the organism as a whole. The same cell state may have completely different functions and meanings in different disease contexts. By closely linking single cell omics data to biological function and disease processes, the state and function of cells can be read more accurately.
Conklin et al. (2022) mentioned that in order to address these challenges, a combination of tools and approaches is needed. The dynamic changes of cell state were tracked by combining the single cell omics data of time series. Technical means such as gene knockout and gene editing can also be used to verify the key role of specific genes or molecules in cell state and function. Learn from the advanced algorithms and technologies in computer science and artificial intelligence to mine the hidden information and rules in the single cell omics data.
4.3 Future direction
Ning et al. (2013) believe that with the continuous development of monocytomics technology, the state and function of cells can be comprehensively understood from different perspectives. In the future, integrating multimodal data will become a critical task. Through the combination of single-cell RNA sequencing, single-cell ATAC sequencing and single-cell proteomics and other single-cell omics technologies, we can more deeply explore the molecular mechanism inside the cell, so as to more comprehensively reveal the physiological and pathological processes of the cell.
Kolodziejczyk et al. (2015) believe that improving spatial resolution is an important direction for future research. If a technology could be developed that could provide high spatial resolution while maintaining single-cell resolution information, it could know exactly where each cell is in the tissue, and how they interact with each other, like a GPS location. This is essential to reveal the mechanisms of immune regulation in the cellular microenvironment. Through such research, more in-depth understanding of the working principle of the immune system, so as to provide new ideas and methods for future immunotherapy.
Of course, to achieve these goals, new tools and algorithms need to be constantly developed. Existing bioinformatics tools and algorithms are often inadequate in the face of large-scale single-cell data. There is a need to develop new tools that are more efficient and accurate to meet the growing data challenges. These new tools will be able to better process and analyze data, improve the accuracy and efficiency of the analysis, and thus further advance the development of immunology research.
4.4 Impact on disease treatment
Single cell omics technology provides a strong support for the realization of personalized medicine. The traditional medical model often adopts a "one-size-fits-all" treatment plan, which lacks sufficient consideration of individual differences. However, single-cell omics technology, through the fine analysis of the patient's specific immune cell status and function, allows doctors to gain insight into the patient's disease characteristics and physiological status, so as to formulate a treatment plan that is more consistent with the individual characteristics of the patient.
Wills and Mead (2015) mention that in cancer treatment, single-cell omics techniques can help physicians analyze the type and number of immune cells in a patient's tumor tissue, as well as their interactions with cancer cells. This helps doctors select the most appropriate immunotherapy approach for the patient, such as personalized immunocell therapy or immunomodulatory drugs, thereby improving the effectiveness and safety of the treatment. The application of single cell omics technology in the field of precision immunotherapy is also of great significance. Traditional immunotherapy approaches often rely on a general understanding of the overall immune system and lack a detailed grasp of the individual immune response. Single-cell omics technology can reveal the complexity and diversity of individual immune responses of patients, providing a strong support for precision immunotherapy.
Macaulay and Voet (2014) suggest that this technique could help identify new immunotherapeutic targets. By analyzing the gene expression profile and protein composition of individual immune cells, researchers can discover key molecules closely related to the development of disease, thus providing new candidate targets for drug development. Single-cell omics technology can also be used to monitor therapeutic effects and guide clinical treatment decisions. By comparing and analyzing the immune cell status of patients before and after receiving immunotherapy, doctors can timely understand the treatment effect and the patient's immune response, so as to adjust the treatment plan or change drugs to ensure the effectiveness and safety of treatment.
Gawad et al. (2016) mentioned that despite the promising application of single-cell omics technology in immune system research and disease treatment, there are still technical and biological challenges. For example, there are still many difficulties in the acquisition and processing of single-cell samples, and the analysis and interpretation of data. However, with the continuous development of new technologies and the application of new algorithms, it is believed that these challenges will be gradually overcome in the future, and single cell omics technology will provide more in-depth insights and more effective solutions for immunological research and clinical treatment.
Bashford-Rogers R., Smith K., and Thomas D., 2018, Antibody repertoire analysis in polygenic autoimmune diseases, Immunology, 155: 3-17.
https://doi.org/10.1111/imm.12927
PMid:29574826 PMCid:PMC6099162
Bettelli E., Carrier Y., Gao W., Korn T., Strom T., Oukka M., Weiner H., and Kuchroo V., 2006, Reciprocal developmental pathways for the generation of pathogenic effector TH17 and regulatory T cells, Nature, 441: 235-238.
https://doi.org/10.1038/nature04753
PMid:16648838
Butler A., Hoffman P., Smibert P., Papalexi E., and Satija R., 2018, Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nature Biotechnology, 36: 411-420.
https://doi.org/10.1038/nbt.4096
PMid:29608179 PMCid:PMC6700744
Chen K.H., Boettiger A., Moffitt J., Wang S.S., and Zhuang X., 2015, Spatially resolved, highly multiplexed RNA profiling in single cells, Science, 348: 6233.
https://doi.org/10.1126/science.aaa6090
PMid:25858977 PMCid:PMC4662681
Conklin K.Y., Zhang B., Knoten A., Diep D., Lake B., Jain S., and Zhang K., 2022, 10X Genomics Single-Nucleus Multiome (RNA+ATAC) Assay for Profiling Adult Human Tissues v1, 2: 18.
https://doi.org/10.17504/protocols.io.b4dqqs5w
Duhen T., Duhen R., Lanzavecchia A., Sallusto F., and Campbell, D., 2012, Functionally distinct subsets of human FOXP3+ Treg cells that phenotypically mirror effector Th cells, Blood, 19: 4430-4440.
https://doi.org/10.1182/blood-2011-11-392324
PMid:22438251 PMCid:PMC3362361
Francisco L.M., Sage P.T., and Sharpe A.H., 2010, The PD-1 pathway in tolerance and autoimmunity
https://doi.org/10.1111/j.1600-065X.2010.00923.x
PMid:20636820 PMCid:PMC2919275
Gawad C., Koh, W., and Quake, S., 2016, Single-cell genome sequencing: current state of the science. Nature Reviews Genetics, 17: 175-188.
https://doi.org/10.1038/nrg.2015.16
PMid:26806412
Giladi A., and Amit I., 2018, Single-Cell Genomics: A Stepping Stone for Future Immunology Discoveries, Cell, 172: 14-21.
https://doi.org/10.1016/j.it.2019.09.004
PMid:31645299
Gomes T., Teichmann S., and Talavera-López C., 2019, Immunology Driven by Large-Scale Single-Cell Sequencing, Trends in Immunology, 40(11):1011-1021.
https://doi.org/10.1038/nmeth.2772
PMid:24363023
Islam S., Zeisel A., Joost S., Manno G.L., Zając P., Kasper M., Lönnerberg P., and Linnarsson S., 2013, Quantitative single-cell RNA-seq with unique molecular identifiers, Nature Methods, 11:163-166.
https://doi.org/10.1038/s12276-020-00499-2
PMid:32929221 PMCid:PMC8080663
Kashima Y., Sakamoto, Y., Kaneko, K., Seki, M., Suzuki, Y., & Suzuki, A., 2020, Single-cell sequencing techniques from individual to multiomics analyses. Experimental & Molecular Medicine, 52, 1419-1427.
https://doi.org/10.1038/s12276-020-00499-2
PMid:32929221 PMCid:PMC8080663
Kazer S., Aicher T., Muema D., Carroll S., Ordovas-Montañes J., Miao V., Tu A., Ziegler C., Nyquist S., Wong E., Ismail N., Dong M., Moodley A., Berger B., Love J., Dong K., Leslie A., Ndhlovu Z., Ndung’u T., Walker B., and Shalek A., 2020, Integrated single-cell analysis of multicellular immune dynamics during hyperacute HIV-1 infection, Nature Medicine, 26: 511 -518.
https://doi.org/10.1038/s41591-020-0799-2
PMid:32251406 PMCid:PMC7237067
Kolodziejczyk A. A., Kim J. K., Svensson V., Marioni J., and Teichmann S., 2015, The technology and biology of single-cell RNA sequencing, Molecular Cell, 58(4): 610-620.
https://doi.org/10.1016/j.molcel.2015.04.005
PMid:26000846
Levine A., Medoza A., Hemmers S., Moltedo B., Niec R., Schizas M., Hoyos B., Putintseva E., Chaudhry A., Dikiy S., Fujisawa S., Chudakov D., Treuting P., and Rudensky A., 2017, Stability and function of regulatory T cells expressing the transcription factor T-bet, Nature, 546: 421-425.
https://doi.org/10.1038/nature22360
PMid:28607488 PMCid:PMC5482236
Luo S., Wang L., Xiao Y., Cao C., Liu Q., and Zhou Y., 2023, Single-Cell RNA-Sequencing Integration Analysis Revealed Immune Cell Heterogeneity in Five Human Autoimmune Diseases, BIO Integration, 5(1): 18-20.
https://doi.org/10.15212/bioi-2023-0012
Macaulay I., and Voet T., 2014, Single Cell Genomics: Advances and Future Perspectives, PLoS Genetics, 10: 27-34.
https://doi.org/10.1371/journal.pgen.1004126
PMid:24497842 PMCid:PMC3907301
Ning L., Liu, G., Li, G., Hou, Y., Tong, Y., & He, J. (2013). Current Challenges in the Bioinformatics of Single Cell Genomics. Frontiers in Oncology, 4: 2-9.
https://doi.org/10.3389/fonc.2014.00007
PMid:24478987 PMCid:PMC3902584
Patruno L., Milite, S., Bergamin, R., Calonaci, N., D’Onofrio, A., Anselmi, F., Antoniotti, M., Graudenzi, A., & Caravagna, G. (2023). A Bayesian method to infer copy number clones from single-cell RNA and ATAC sequencing. PLOS Computational Biology, 19.
https://doi.org/10.1371/journal.pcbi.1011557
PMid:37917660 PMCid:PMC10645363
Peng Y.M., Luo X.M., and Chen J.Y., 2023, Research Progress in the metabolism of Tumor Microenvironment and New Strategies of Immunotherapy, Journal of Sichuan University (Medical Edition), 54(3): 505-509.
Proserpio V., and Mahata B., 2015, Single-cell technologies to study the immune system, 147(2): 133-140.
https://doi.org/10.1111/imm.12553
PMid:26551575 PMCid:PMC4717243
Stubbington M., Rozenblatt-Rosen O., Regev A., and Teichmann S., 2017, Single-cell transcriptomics to explore the immune system in health and disease, Science, 358: 58-63.
https://doi.org/10.1126/science.aan6828
PMid:28983043 PMCid:PMC5654495
Tanay A., & Regev, A., 2017, Scaling single-cell genomics from phenomenology to mechanism. Nature, 541: 331-338.
https://doi.org/10.1038/nature21350
PMid:28102262 PMCid:PMC5438464
Vegh P., and Haniffa M., 2018, The impact of single-cell RNA sequencing on understanding the functional organization of the immune system, Briefings in Functional Genomics, 17: 265-272.
https://doi.org/10.1093/bfgp/ely003
PMid:29547972 PMCid:PMC6063276
Wang, Y., & Navin, N. (2015). Advances and applications of single-cell sequencing technologies. Molecular Cell, 58(4): 598-609.
https://doi.org/10.1016/j.molcel.2015.05.005
PMid:26000845 PMCid:PMC4441954
Wills Q., & Mead, A. (2015). Application of single-cell genomics in cancer: promise and challenges. Human Molecular Genetics, 24: R74-R84.
https://doi.org/10.1093/hmg/ddv235
PMid:26113645 PMCid:PMC4571998
Wimmers F., Donato, M., Kuo, A., Ashuach, T., Gupta, S., Li, C., Dvorak, M., Foecke, M., Chang, S., Hagan, T., Jong, S., Maecker, H., Most, R., Cheung, P., Cortese, M., Bosinger, S., Davis, M., Rouphael, N., Subramaniam, S., Yosef, N., Utz, P., Khatri, P., & Pulendran, B. (2021). The single-cell epigenomic and transcriptional landscape of immunity to influenza vaccination. Cell, 184: 3915-3935.
https://doi.org/10.1016/j.cell.2021.05.039
PMid:34174187 PMCid:PMC8316438
Yegorov O., Yang C., Rahman M., Ghiaseddin A., Dechkovskaia A., Cleaver B., Hodik M., Amdur R., Massini T., Kresak J., and Mitchell D., 2019, ATIM-34. single-cell rna sequencing reveals dynamic immune response changes in glioblastoma patient with durable complete response to cmv pp65-lamp rna-pulsed dendritic vaccines, Neuro-Oncology, 21(Suppl 6): vi9.
https://doi.org/10.1093/neuonc/noz175.033
PMCid:PMC6847930
Yost K., Chang H., and Satpathy A., 2019, Tracking the immune response with single-cell genomics, Vaccine, 38(28): 4487-4490.
https://doi.org/10.1016/j.vaccine.2019.11.035
PMid:31859202 PMCid:PMC7263961
Yuan S.Y., Xu H., and Xie J.P., 2017, New progress in the efficacy relationship between immune cells and PD-1 and EGFR-TKIs in the tumor microenvironment, Chinese Journal of Lung Cancer, 20(11): 775-780.
. PDF(386KB)
. FPDF(win)
. FPDF(mac)
. HTML
. Online fPDF
Associated material
. Readers' comments
Other articles by authors
. Wang Wei
Related articles
. Single-cell omics
. Immune system dynamics
. Heterogeneity analysis
. Precision immunotherapy
Tools
. Email to a friend
. Post a comment